I started writing this post in the train from Norrköpping back to Stockholm Arlanda airport after visiting the truly excellent EG VCBM 2012 (Eurographics Workshop on Visual Computing for Biology and Medicine). While enjoying the beautiful view on the stunning Swedish landscape that features many lakes, trees with autumn leaves and a nice autumn sun, I briefly summarized the conference highlights. Please note that these were the conference highlights for me personally, and are not necessarily a reflection of the actual conference highlights. There were so many great talks, I lost count, but in the interest of not making this blog post drag on for too long, I’ll restrict myself to just briefly summarizing a couple of them here.
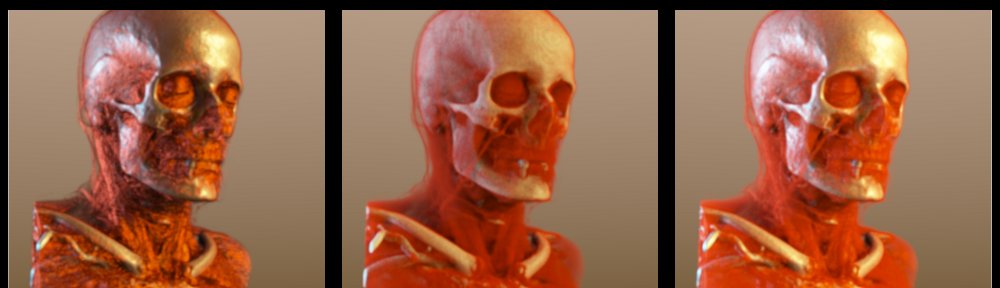
The first day started with a word of welcome from the chairs Timo Ropinski and Anders Ynnerman and an excellent keynote by Anders Persson (Director of the CMIV): ‘Visualization of Quantified Medical Image Data – Key to the Future?’. While showing us beautiful datasets acquired from Dual Energy CT (DECT), he stressed the importance of making quantified imaging data usable in clinical practice. This can be achieved by working in close collaboration with medical centers to make sure the techniques we are developing are usable by and useful to clinicians or medical researchers.
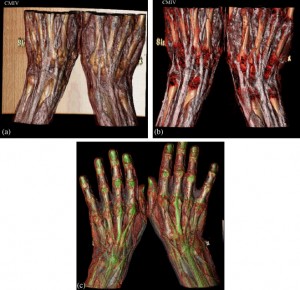
Dual-energy CT (DECT) with two X-ray sources running simultaneously at different energies allows obtaining additional information about the elementary chemical composition of computer tomography scanned material [1].
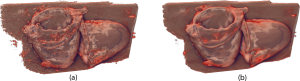
Raw 3D ultrasound scan on the left and a visualization of the same dataset filtered with the lowest-variance streamline method [4].
On the second conference day, the Segmentation and Simulation session’s second talk presented by Frank Heckel ‘Sketch-based Image-Independent Editing of 3D Tumor Segmentations Using Variational Interpolation’ was also very interesting [6]. By allowing the user to intuitively draw adjustments on single slices, automatic segmentations can easily be adjusted by medical experts. This is currently shown on CT, but since the adjustments are not image-based, the technique is valid for arbitrary modalities.
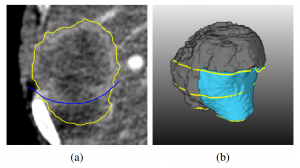
Sketch-based segmentation editing: (a) initial segmentation (yellow), manual correction (blue) and (b) 3D result after editing with our variational-interpolation-based approach [6].
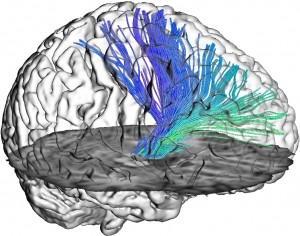
The medvis.org overlord who also happens to be my supervisor, Charl Botha, presented ‘BrainCove: A Tool for Voxel-Wise fMRI Brain Connectivity Visualization’ in the same session [8]. I am obviously biased here, but I thought it was an interesting and entertaining presentation. I mean it featured the BrainCleaver, what’s not to like?
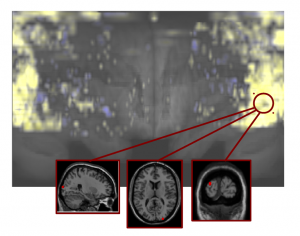
Braincove’s Lambert’s Cylindrical flatmap representation
of the brain, viewing from the anterior in the middle to posterior at the two sides [8].
References
- [1] Anders Persson, Christian Jackowskia, Elias Engström and Helene Zachrisson, “Advances of dual source, dual-energy imaging in postmortem CT”, European Journal of Radiology, Volume 68, Issue 3, December 2008, Pages 446–455
- [2]: Anne Berres, Mathias Goldau, Marc Tittgemeyer, Gerik Scheuermann and Hans Hagen, “Tractography in Context: Multimodal Visualization of Probabilistic Tractograms in Anatomical Context”
- [3]: D. Tenbrinck, A. Sawatzky, X. Jiang, M. Burger, W. Haffner, P. Willems, M. Paul and J. Stypmann “Impact of Physical Noise Modeling on Image Segmentation in Echocardiography”
- [4]: Veronika Šoltészová, Linn Emilie Sævil Helljesen, Wolfgang Wein, Odd Helge Gilja and Ivan Viola, “Lowest-Variance Streamlines for Filtering of 3D Ultrasound”
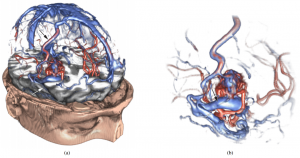
- [5]: F. Weiler, C. Rieder, C. A. David, C. Wald, and H. K. Hahn “On the Value of Multi-Volume Visualization for Preoperative Planning of Cerebral AVM Surgery”
- [6]: F. Heckel, S. Braunewell, G. Soza, C. Tietjen and H. K. Hahn, “Sketch-based Image-independent Editing of 3D Tumor Segmentations using Variational Interpolation”
- [7]: Tobias Moench, Christoph Kubisch, Kai Lawonn, Ruediger Westermann and Bernhard Preim,”Visually Guided Mesh Smoothing for Medical Applications”
- [8]: A.F. van Dixhoorn, J. Milles, B. van Lew and C.P. Botha “BrainCove: A Tool for Voxel-wise fMRI Brain Connectivity Visualization”
- [9]: Katja Mogalle, Christian Tietjen, Grzegorz Soza and Bernhard Preim, “Constrained Labeling of 2D Slice Data for Reading Images in Radiology”
Pingback: A look back on 2012 | Noeska Smit